Rainfall as a triggering factor in a spatial domain#
© 2024 Daniel F. Ruiz, Exneyder A. Montoya-Araque y Universidad EAFIT.
This notebook can be interactively run in Google - Colab.
This notebook runs the model TRIGRS (Transient Rainfall Infiltration and Grid-Based Regional Slope-Stability Analysis), developed by Baum et al. [2002] and Baum et al. [2008], and parallelized later by Alvioli and Baum [2016]
Required modules and global setup for plots#
import os
import subprocess
import textwrap
import itertools
import requests
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib as mpl
from IPython import get_ipython
if 'google.colab' in str(get_ipython()):
print('Running on CoLab. Installing the required modules...')
# subprocess.run('pip install ipympl', shell=True);
subprocess.run('pip install palettable', shell=True);
subprocess.run('pip install pynewmarkdisp', shell=True);
from google.colab import output, files
output.enable_custom_widget_manager()
else:
import tkinter as tk
from tkinter.filedialog import askopenfilename
from pynewmarkdisp.spatial import get_hillshade
# Figures setup
# %matplotlib widget
%matplotlib inline
mpl.rcParams.update({
"font.family": "serif",
"font.serif": ["Computer Modern Roman", "cmr", "cmr10", "DejaVu Serif"], # or
"mathtext.fontset": "cm", # Use Computer Modern fonts for math
"axes.formatter.use_mathtext": True, # Use mathtext for axis labels
"axes.unicode_minus": False, # Use standard minus sign instead of a unicode character
})
Creating directories#
# Create a folder called TRIGRS in the current working directory if it doesn't exist
workdir = os.getcwd()
exec_dir = os.path.join(workdir, "TRIGRS_2")
os.makedirs(f"{exec_dir}", exist_ok=True)
# Create a folder inside TRIGRS to store the ascii files
inputs_dir = os.path.join(exec_dir, "inputs")
outputs_dir = os.path.join(exec_dir, "outputs")
os.makedirs(f"{inputs_dir}", exist_ok=True)
os.makedirs(f"{outputs_dir}", exist_ok=True)
Functions#
def tpx_in_maker(tpx_inputs):
tpx_in_template = textwrap.dedent(
f"""\
TopoIndex 1.0.15; Name of project (up to 255 characters)
TopoIndex analysis - Project: {tpx_inputs["proj_name"]}
Flow-direction numbering scheme (ESRI=1, TopoIndex=2)
{tpx_inputs["flow_dir_scheme"]}
Exponent, Number of iterations
{tpx_inputs["exponent_weight_fact"]}, {tpx_inputs["iterarions"]}
Name of elevation grid file
TRIGRS_2/inputs/dem.asc
Name of direction grid
TRIGRS_2/inputs/directions.asc
Save listing of D8 downslope neighbor cells (TIdsneiList_XYZ)? Enter T (.true.) or F (.false.)
T
Save grid of D8 downslope neighbor cells (***TIdscelGrid_XYZ)? Enter T (.true.) or F (.false.)
T
Save cell index number grid (TIcelindxGrid_XYZ)? Enter T (.true.) or F (.false.)
T
Save list of cell number and corresponding index number (***TIcelindxList_XYZ)? Enter T (.true.) or F (.false.)
T
Save flow-direction grid remapped from ESRI to TopoIndex (TIflodirGrid_XYZ)? Enter T (.true.) or F (.false.)
T
Save grid of points on ridge crests (TIdsneiList_XYZ)? Enter T (.true.) or F (.false.); Sparse (T) or dense (F)?
F,T
ID code for output files? (8 characters or less)
{tpx_inputs["proj_name"]}"""
)
with open("tpx_in.txt", "w") as file:
file.write(tpx_in_template)
return
def run_topoidx(tpx_inputs):
tpx_in_maker(tpx_inputs) # Generating initializing files
subprocess.call(["chmod +x ./TRIGRS_2/Exe_TopoIndex"], shell=True) # Change permission
subprocess.call(["./TRIGRS_2/Exe_TopoIndex"]) # Running Fortran executable
# Moving log file to outputs folder and renaming it
os.rename(os.path.join(workdir, "TopoIndexLog.txt"), os.path.join(outputs_dir, f"TopoIndexLog_{tpx_inputs['proj_name']}.txt"))
os.rename(os.path.join(workdir, "tpx_in.txt"), os.path.join(exec_dir, f"tpx_in_{tpx_inputs['proj_name']}.txt"))
return
def tr_in_maker(trg_inputs):
# Create text block for geotechnical parameters of each zone
geoparams = ""
for zone in trg_inputs["geoparams"].keys():
z = trg_inputs["geoparams"][zone]
txt_zone = "".join(
[
f"zone, {zone}\n",
"cohesion, phi, uws, diffus, K-sat, Theta-sat, Theta-res, Alpha\n",
f'{z["c"]}, {z["φ"]}, {z["γ_sat"]}, {z["D_sat"]}, {z["K_sat"]}, {z["θ_sat"]}, {z["θ_res"]}, {z["α"]}\n',
]
)
# print(txt_zone)
geoparams = str().join([geoparams, txt_zone])
# Update rifil entry with placeholder for each file
trg_inputs["rifil"] = str().join(
[f"TRIGRS_2/inputs/ri{n}.asc\n" for n in range(1, len(trg_inputs["capt"]))]
)
# Template
trg_in_template = (
textwrap.dedent(
f"""\
Project: {trg_inputs['proj_name']}
TRIGRS, version 2.1.00c, (meter-kilogram-second-degrees units)
tx, nmax, mmax, zones
{trg_inputs['tx']}, {trg_inputs['nmax']}, {trg_inputs['mmax']}, {trg_inputs['zones']}
nzs, zmin, uww, nper t
{trg_inputs['nzs']}, 0.001, 9.8e3, {trg_inputs['nper']}, {trg_inputs['t']}
zmax, depth, rizero, Min_Slope_Angle (degrees), Max_Slope_Angle (degrees)
{trg_inputs['zmax']}, {trg_inputs['depth']}, {trg_inputs['rizero']}, 0., 90.0
"""
)
+ geoparams[:-1]
+ textwrap.dedent(
f"""
cri(1), cri(2), ..., cri(nper)
{', '.join(map(str, trg_inputs['cri']))}
capt(1), capt(2), ..., capt(n), capt(n+1)
{', '.join(map(str, trg_inputs['capt']))}
File name of slope angle grid (slofil)
TRIGRS_2/inputs/slope.asc
File name of digital elevation grid (elevfil)
TRIGRS_2/inputs/dem.asc
File name of property zone grid (zonfil)
TRIGRS_2/inputs/zones.asc
File name of depth grid (zfil)
TRIGRS_2/inputs/zmax.asc
File name of initial depth of water table grid (depfil)
TRIGRS_2/inputs/depthwt.asc
File name of initial infiltration rate grid (rizerofil)
TRIGRS_2/inputs/rizero.asc
List of file name(s) of rainfall intensity for each period, (rifil())
"""
)
+ trg_inputs["rifil"][:-2]
+ textwrap.dedent(
f"""
File name of grid of D8 runoff receptor cell numbers (nxtfil)
TRIGRS_2/inputs/TIdscelGrid_{trg_inputs['proj_name']}.txt
File name of list of defining runoff computation order (ndxfil)
TRIGRS_2/inputs/TIcelindxList_{trg_inputs['proj_name']}.txt
File name of list of all runoff receptor cells (dscfil)
TRIGRS_2/inputs/TIdscelList_{trg_inputs['proj_name']}.txt
File name of list of runoff weighting factors (wffil)
TRIGRS_2/inputs/TIwfactorList_{trg_inputs['proj_name']}.txt
Folder where output grid files will be stored (folder)
TRIGRS_2/outputs/
Identification code to be added to names of output files (suffix)
{trg_inputs['proj_name']}
Save grid files of runoff? Enter T (.true.) or F (.false.)
F
Save grid of minimum factor of safety? Enter T (.true.) or F (.false.)
T
Save grid of depth of minimum factor of safety? Enter T (.true.) or F (.false.)
T
Save grid of pressure head at depth of minimum factor of safety? Enter T (.true.) or F (.false.)
T
Save grid of computed water table depth or elevation? Enter T (.true.) or F (.false.) followed by 'depth,' or 'eleva'
T, depth
Save grid files of actual infiltration rate? Enter T (.true.) or F (.false.)
F
Save grid files of unsaturated zone basal flux? Enter T (.true.) or F (.false.)
F
Save listing of pressure head and factor of safety ("flag")? (-9 sparse xmdv , -8 down-sampled xmdv, -7 full xmdv, -6 sparse ijz, -5 down-sampled ijz, -4 full ijz, -3 Z-P-Fs-saturation list -2 detailed Z-P-Fs, -1 Z-P-Fs list, 0 none). Enter flag value followed by down-sampling interval (integer).
{trg_inputs['ψ_flag']}, {trg_inputs['nzs']}
Number of times to save output grids and (or) ijz/xmdv files
{trg_inputs['n_outputs']}
Times of output grids and (or) ijz/xmdv files
{', '.join(map(str, trg_inputs['t_n_outputs']))}
Skip other timesteps? Enter T (.true.) or F (.false.)
F
Use analytic solution for fillable porosity? Enter T (.true.) or F (.false.)
T
Estimate positive pressure head in rising water table zone (i.e. in lower part of unsat zone)? Enter T (.true.) or F (.false.)
T
Use psi0=-1/alpha? Enter T (.true.) or F (.false.) (False selects the default value, psi0=0)
F
Log mass balance results? Enter T (.true.) or F (.false.)
T
Flow direction (Enter "gener", "slope", or "hydro")
{trg_inputs['flowdir']}
Add steady background flux to transient infiltration rate to prevent drying beyond the initial conditions during periods of zero infiltration?
T
Specify file extension for output grids. Enter T (.true.) for ".asc" or F for ".txt"
T
Ignore negative pressure head in computing factor of safety (saturated infiltration only)? Enter T (.true.) or F (.false.)
T
Ignore height of capillary fringe in computing pressure head for unsaturated infiltration option? Enter T (.true.) or F (.false.)
T
Parameters for deep pore-pressure estimate in SCOOPS ijz output: Depth below ground surface (positive, use negative value to cancel this option), pressure option (enter 'zero' , 'flow' , 'hydr' , or 'relh')
-50.0,flow
"""
)
)
# Create txt file
with open("tr_in.txt", "w") as file:
file.write(trg_in_template)
return
def run_trigrs(trg_inputs):
# Correcting sign for `Alpha` parameters for saturated/unsaturated models
for z in trg_inputs["geoparams"].keys():
zone = trg_inputs["geoparams"][z]
if "S" in trg_inputs["model"]:
zone["α"] = -1 * abs(zone["α"])
else:
zone["α"] = abs(zone["α"])
# Correcting sign for `mmax` parameters for finite/infinite models
if "I" in trg_inputs["model"]:
trg_inputs["mmax"] = -1 * abs(trg_inputs["mmax"])
else:
trg_inputs["mmax"] = abs(trg_inputs["mmax"])
# Generating initializing file and running TRIGRS executable
tr_in_maker(trg_inputs) # Initializing files from template
if trg_inputs["parallel"]:
exe = "./TRIGRS_2/Exe_TRIGRS_par"
subprocess.call([f"chmod +x {exe}"], shell=True) # Change permission
subprocess.call(
[f'mpirun -np {trg_inputs["NP"]} {exe}'], shell=True
) # Running TRIGRS executable
else:
exe = "./TRIGRS_2/Exe_TRIGRS_ser"
subprocess.call([f"chmod +x {exe}"], shell=True) # Change permission
subprocess.call([exe], shell=True) # Running TRIGRS executable
# Moving files to output folders and renaming it
proj_name, model = trg_inputs["proj_name"], trg_inputs["model"]
os.rename(os.path.join(workdir, "TrigrsLog.txt"), os.path.join(outputs_dir, f"TrigrsLog_{proj_name}_{model}.txt"))
os.rename(os.path.join(workdir, "tr_in.txt"), os.path.join(exec_dir, f"tr_in_{proj_name}_{model}.txt"))
groups = (
"TRfs_min",
"TRp_at_fs_min",
"TRwater_depth",
"TRz_at_fs_min",
"TR_ijz_p_th",
# "TRrunoffPer",
)
[os.makedirs(f"TRIGRS_2/outputs/{grp}", exist_ok=True) for grp in groups]
idx = np.arange(1, trg_inputs["n_outputs"] + 1, 1)
for grp, i in itertools.product(groups, idx):
if grp == "TRrunoffPer":
os.rename(
f"TRIGRS_2/outputs/{grp}{i}{proj_name}.asc",
f"TRIGRS_2/outputs/{grp}/{grp}_{proj_name}_{model}_{i}.asc",
)
elif grp == "TR_ijz_p_th":
os.rename(
f"TRIGRS_2/outputs/{grp}_{proj_name}_{i}.txt",
f"TRIGRS_2/outputs/{grp}/{grp}_{proj_name}_{model}_{i}.txt",
)
else:
os.rename(
f"TRIGRS_2/outputs/{grp}_{proj_name}_{i}.asc",
f"TRIGRS_2/outputs/{grp}/{grp}_{proj_name}_{model}_{i}.asc",
)
return
def get_param(zone, param):
return trg_inputs['geoparams'][zone.iloc[0]][param]
def get_df(file):
γ_w = 9.8e3 # [N/m3] unit weight of water
# Reading file
df = pd.read_csv(file,
names=['i_tr', 'j_tr', 'elev', 'ψ', 'θ'],
skiprows=7, sep='\s+')#delim_whitespace=True)
# Correcting indices to match numpy notation
df['i'] = df['j_tr'].max() - df['j_tr']
df['j'] = df['i_tr'] - 1
df.drop(columns=['i_tr', 'j_tr'], inplace=True)
# Calculating depths from elevations
elev_max = df.groupby(['i', 'j'])['elev'].transform('max')
df['z'] = elev_max - df['elev'] + 0.001
df = df[['i', 'j', 'elev', 'z', 'ψ', 'θ']]
# Assigning zones and geotechnical parameters
df['zone'] = zones[df['i'], df['j']]
for param_name in trg_inputs['geoparams'][1].keys():
df[param_name] = df.groupby('zone')['zone'].transform(
get_param, param=param_name)
# Correting rows with ψ==0 (watertable) due to numerical errors
mask = (df['ψ'] == 0) & (df['z'] > 0.001) & (df['θ'] == df['θ_sat'])
# df.loc[mask, 'z'] = df.loc[mask, 'z'] + 1/df.loc[mask, 'α']
# df.loc[mask, 'elev'] = df.loc[mask, 'elev'] - 1/df.loc[mask, 'α']
# df.sort_values(['i', 'j', 'z'], inplace=True)
df.drop(df[mask].index, inplace=True) # This eliminates the wt rows
# Assigning slope
df['δ'] = slope[df['i'], df['j']]
δrad = np.deg2rad(df['δ'])
# Calculating effective stress parameter, χ
df['χ'] = (df['θ'] - df['θ_res']) / (df['θ_sat'] - df['θ_res'])
# Specific gravity
df['Gs'] = (df['γ_sat']/γ_w - df['θ_sat']) / (1 - df['θ_sat'])
# unit weight at unsat and sat zones
df['γ_nat'] = (df['Gs'] * (1 - df['θ_sat']) + df['θ']) * γ_w
# average unit weight
γ_cumul = df.groupby(['i', 'j'])['γ_nat'].transform('cumsum')
idx = df.groupby(['i', 'j'])['γ_nat'].cumcount() + 1
df['γ_avg'] = γ_cumul / idx
# Factor of safety
fs1 = np.tan(np.deg2rad(df['φ'])) / np.tan(δrad)
fs2_num = df['c'] - df['ψ'] * γ_w * df['χ'] * np.tan(np.deg2rad(df['φ']))
fs2_den = df['γ_avg'] * df['z'] * np.sin(δrad) * np.cos(δrad)
fs = fs1 + fs2_num/fs2_den
fs[fs > 3] = 3
df['fs'] = fs
return df
Loading executables and input files#
def download_file_from_github(file, url_path, output_path, make_executable=False):
os.makedirs(output_path, exist_ok=True)
destination = os.path.join(output_path, file)
# ✅ Check if file already exists
if os.path.exists(destination):
print(f"🟡 File already exists at: {destination} — skipping download.")
return
url = f"{url_path.rstrip('/')}/{file}"
try:
print(f"⬇️ Downloading from: {url}")
subprocess.run(["wget", "-q", "-O", destination, url], check=True)
if make_executable:
os.chmod(destination, 0o755)
print(f"🔧 File marked as executable.")
print(f"✅ File downloaded to: {destination}")
except subprocess.CalledProcessError as e:
print(f"❌ Download failed: {e}")
url_path = "https://raw.githubusercontent.com/eamontoyaa/data4testing/main/trigrs/" # URL of the raw file on GitHub
output_path = "./TRIGRS_2/" # Path where you want to save the downloaded file
TopoIndex#
file = "Exe_TopoIndex" # Name of the file to download
download_file_from_github(file, url_path, output_path)
⬇️ Downloading from: https://raw.githubusercontent.com/eamontoyaa/data4testing/main/trigrs/Exe_TopoIndex
✅ File downloaded to: ./TRIGRS_2/Exe_TopoIndex
TRIGRS#
file = "Exe_TRIGRS_ser" # Name of the file to download
download_file_from_github(file, url_path, output_path)
⬇️ Downloading from: https://raw.githubusercontent.com/eamontoyaa/data4testing/main/trigrs/Exe_TRIGRS_ser
✅ File downloaded to: ./TRIGRS_2/Exe_TRIGRS_ser
# Check the files in the executables folder
os.listdir(exec_dir)
['Exe_TopoIndex', 'outputs', 'Exe_TRIGRS_ser', 'inputs']
Running the TRIGRS model for a one-cell spatial domain#
General inputs#
proj_name = "SPATIAL"
model = "SI" # Choose among UF, UI, SF, SI → | U: unsaturated | S: saturated | F: finite | I: infinite |
rainfall_int = np.array([3.e-7, 9.e-5]) # [m/s]
# rainfall_int = np.array([0.5e-4, 0.5e-4]) # [m/s]
cumul_duration = np.array([0, 172800, 216000]) # [s] Always starts at 0 and contains n+1 elements, where n is the number of rainfall intensities values
output_times = np.array([0.01, 43200, 86400, 100800, 183600, 216000]) # [s] It does not have to include the same values as cumul_duration
Creating .asc files#
def load_ascii_raster(path, dtype=None):
raster = np.loadtxt(path, skiprows=6)
header = np.loadtxt(path, max_rows=6, dtype=object)
header = {
"ncols": int(header[0, 1]),
"nrows": int(header[1, 1]),
"xllcorner": float(header[2, 1]),
"yllcorner": float(header[3, 1]),
"cellsize": float(header[4, 1]),
"nodata_value": eval(header[5, 1]),
}
raster[np.where(raster == header["nodata_value"])] = np.nan
raster = raster.astype(dtype) if dtype is not None else raster
return (raster, header)
def get_asc_from_url(file_name):
base_url = "https://raw.githubusercontent.com/eamontoyaa/data4testing/main/trigrs/tutorial_data"
url = f"{base_url}/{file_name}.asc"
output_path = os.path.join(inputs_dir, f"{file_name}.asc")
if os.path.exists(output_path):
print(f"🟡 {file_name}.asc already exists at {output_path} — skipping download.")
return
try:
subprocess.run(["wget", "-q", "-O", output_path, url], check=True)
print(f"✅ {file_name}.asc downloaded successfully.")
except subprocess.CalledProcessError:
print(f"❌ Failed to download {file_name}.asc from {url}")
for file in ['dem', 'zmax', 'depthwt', 'slope', 'directions', 'rizero', 'zones']:
get_asc_from_url(file)
✅ dem.asc downloaded successfully.
✅ zmax.asc downloaded successfully.
✅ depthwt.asc downloaded successfully.
✅ slope.asc downloaded successfully.
✅ directions.asc downloaded successfully.
✅ rizero.asc downloaded successfully.
✅ zones.asc downloaded successfully.
Note:
If you are loading locally your own spatial input data, rerun the notebook from this point.
dem, header = load_ascii_raster(f"{inputs_dir}/dem.asc")
slope, header = load_ascii_raster(f"{inputs_dir}/slope.asc")
directions, header = load_ascii_raster(f"{inputs_dir}/directions.asc")
zones, header = load_ascii_raster(f"{inputs_dir}/zones.asc")
depth, header = load_ascii_raster(f"{inputs_dir}/zmax.asc")
depth, header = load_ascii_raster(f"{inputs_dir}/rizero.asc")
depth_w, header = load_ascii_raster(f"{inputs_dir}/depthwt.asc")
# Running TopoIndex
tpx_inputs = {
"flow_dir_scheme": 2,
"exponent_weight_fact": 25,
"iterarions": 10,
"proj_name": proj_name,
}
run_topoidx(tpx_inputs)
TopoIndex: Topographic Indexing and
flow distribution factors for routing
runoff through Digital Elevation Models
By Rex L. Baum
U.S. Geological Survey
Version 1.0.14, 11May2015
-----------------------------------------
TopoIndex analysis - Project: SPATIAL
100 = number of data cells
100 = total number of cells
Reading elevation grid data
Initial elevation indexing completed
Finding D8 neighbor cells
nxtcel() nodata (integer,floating)= -9999 -9999.00000
Identifying downslope cells and grid mismatches
No grid mismatch found!
Correcting cell index numbers
Computing weighting factors
Saving results to disk
trg_inputs = {
"proj_name": proj_name, # Name of the project
"model": model, # S-saturated or U-unsaturated + F-finite or I-infinite
"tx": 20, # [] tx*nper determines how many time steps are used in the computations
"nmax": 30, # [] Ma number of roots 𝚲n and terms in series solutions for UNS.zone
"mmax": 100, # [] Max number of terms in series solution for FIN depth. If <0: INF depth
"zones": 2, # [] Number of zones
"nzs": 20, # [] Vertical increments until zmax
"nper": len(rainfall_int), # [-] Periods of rainfall (N)
"t": cumul_duration[-1], # [s] Elapsed time since the start of the storm (t)
"zmax": -3.001, # [m] Max. depth to compute Fs and Ψ (if <0 read from grid file)
"depth": -2.4, # [m] Water table depth (d) (if <0 read from grid file)
"rizero": -1e-9, # [m/s] Steady pre-storm and long-term infilt. (Izlt) (if <0 read from grid file)
"geoparams":{
1: {
"c": 3.5e3, # [Pa] soil cohesion for effective stress (c')
"φ": 35, # [deg] soil friction angle for effective stress (φ')
"γ_sat": 22.0e3, # [N/m3] unit weight of soil (γs)
"D_sat": 6.0e-6, # [m2/s] Saturated hydraulic diffusivity (D0)
"K_sat": 1.0e-7, # [m/s] Saturated hydraulic conductivity (Ks)
"θ_sat": 0.45, # [-] Saturated water content (θs)
"θ_res": 0.05, # [-] Residual water content (θr)
"α": 0.5, # [1/m] Fitting parameter. If <0: saturated infiltration
},
2: {
"c": 8e3, # [Pa] soil cohesion for effective stress (c')
"φ": 31, # [deg] soil friction angle for effective stress (φ')
"γ_sat": 22.0e3, # [N/m3] unit weight of soil (γs)
"D_sat": 8.0e-4, # [m2/s] Saturated hydraulic diffusivity (D0)
"K_sat": 1.0e-4, # [m/s] Saturated hydraulic conductivity (Ks)
"θ_sat": 0.45, # [-] Saturated water content (θs)
"θ_res": 0.06, # [-] Residual water content (θr)
"α": 8, # [1/m] Fitting parameter. If <0: saturated infiltration
}
},
"cri": tuple(rainfall_int), # [m/s] Rainfall intensities (Inz)
"capt": tuple(cumul_duration), # [s] Cumulative duration of rainfall cri[i]
"flowdir": "gener", # Method for maximum allowed Ψ ("gener", "slope", or "hydro"))
"n_outputs": len(output_times), # [-] No of raster outputs at different t
"t_n_outputs": tuple(output_times),#t, # [s] Time at which to save raster files
"ψ_flag": -4,
"parallel": False, # [-] Run in parallel
"NP" : 4, # Number of processors
# "inz_n_outputs": inz_n_outputs #inz, # [s] Rainfall intensities at which to save raster files
}
run_trigrs(trg_inputs)
TRIGRS: Transient Rainfall Infiltration
and Grid-based Regional Slope-Stability
Analysis
Version 2.1.00c, 02 Feb 2022
By Rex L. Baum and William Z. Savage
U.S. Geological Survey
-----------------------------------------
Opening default initialization file
TRIGRS, version 2.1.00c, (meter-kilogram-second-degrees units)
Negative or zero value of Alpha for property zone 1
Saturated infiltration model will be used for cells in zone 1 .
Negative or zero value of Alpha for property zone 2
Saturated infiltration model will be used for cells in zone 2 .
******** Zone 1 *********
Using saturated infiltration model to avoid
early-time errors in unsaturated infiltration model.
******** Zone 2 *********
Using saturated infiltration model to avoid
early-time errors in unsaturated infiltration model.
******** ******** ******** *********
TRIGRS_2/inputs/TIgrid_size.txt T
Reading input grids
Testing and adjusting steady infiltration rates
Adjusted steady infiltration rate at 0 cells
Starting runoff-routing computations
Initial size of time-steps 5400.0000000000000
******** Output times ********
number, timestep #, time
One or more specified output times unavailable,
Nearest available time substituted.
1 1 0.00000000
One or more specified output times unavailable,
Nearest available time substituted.
2 6 43200.0000
One or more specified output times unavailable,
Nearest available time substituted.
3 11 86400.0000
One or more specified output times unavailable,
Nearest available time substituted.
4 12 95040.0000
One or more specified output times unavailable,
Nearest available time substituted.
5 26 183600.000
One or more specified output times unavailable,
Nearest available time substituted.
6 41 216000.000
Starting computations of pressure head and factor of safety
Ignoring unsaturated zone
Calling multistep saturated infinite-depth model
Starting computations
for infinite-depth saturated zone
Cells completed:
0
100 cells completed
Saving results
TRIGRS finished!
STOP 0
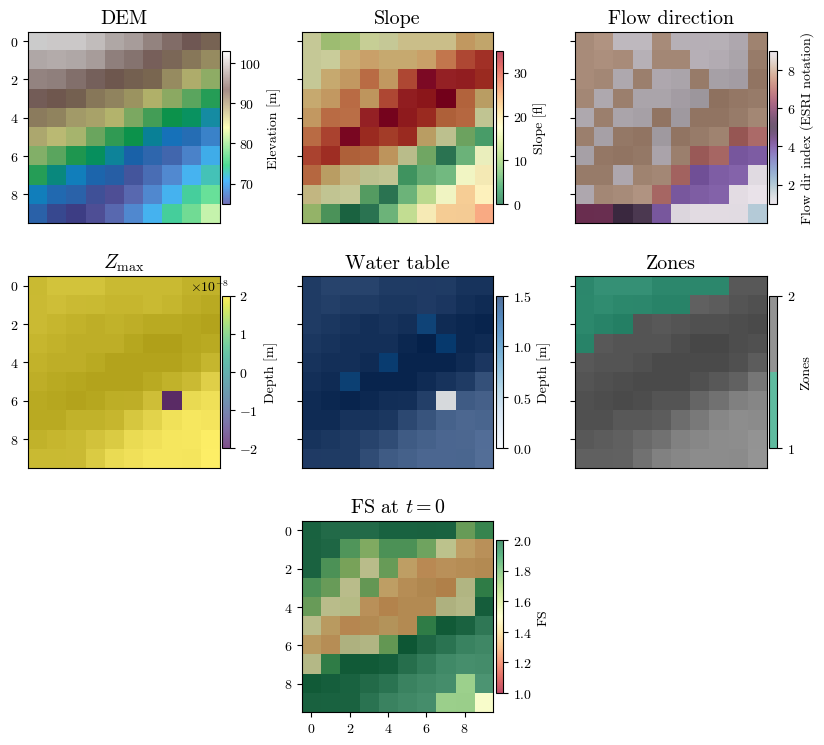
Visualizing spatial inputs (conditioning factors)#
hsd = get_hillshade(dem, sun_azimuth=000, sun_altitude=30, cellsize=header['cellsize'])
fig, axs = plt.subplot_mosaic([['a', 'b', 'c'], ['d', 'e', 'f'], ['.', 'g', '.']],
figsize=(10, 3*3), sharex=True, sharey=True)
(ax0, ax1, ax2, ax3, ax4, ax5, ax6) = axs.values()
for ax in [ax0, ax1, ax2, ax3, ax4, ax5, ax6]:
im_hsd = ax.matshow(hsd, cmap="gray", interpolation="none")
# DEM
im_dem = ax0.matshow(dem, cmap="terrain", alpha=0.7, interpolation="none")
ax0.set_title("DEM", fontsize='x-large')
fig.colorbar(im_dem, ax=ax0, orientation='vertical', shrink=0.75, pad=0.01, label='Elevation [m]')
# Slope
im_slope = ax1.matshow(slope, cmap="RdYlGn_r", alpha=0.7, interpolation="none")
ax1.set_title("Slope", fontsize='x-large')
fig.colorbar(im_slope, ax=ax1, orientation='vertical', shrink=0.75, pad=0.01, label='Slope [°]')
# Directions
im_dirflow = ax2.matshow(directions, cmap="twilight", alpha=0.7, interpolation="none")
ax2.set_title("Flow direction", fontsize='x-large')
fig.colorbar(im_dirflow, ax=ax2, orientation='vertical', shrink=0.75, pad=0.01, label='Flow dir index (ESRI notation)')
# Zmax
im_zmax = ax3.matshow(depth, cmap='viridis', alpha=0.7, interpolation="none")
ax3.set_title("$Z_\\mathrm{max}$", fontsize='x-large')
fig.colorbar(im_zmax, ax=ax3, orientation='vertical', shrink=0.75, pad=0.01, label='Depth [m]')
# Water table depth
im_depth = ax4.matshow(depth_w, cmap='Blues', alpha=0.7, interpolation="none")
ax4.set_title("Water table", fontsize='x-large')
fig.colorbar(im_depth, ax=ax4, orientation='vertical', shrink=0.75, pad=0.01, label='Depth [m]')
# Zones
im_zones = ax5.matshow(zones, cmap=plt.colormaps.get_cmap("Dark2").resampled(len(np.unique(zones))),
alpha=0.7, interpolation="none")
ax5.set_title("Zones", fontsize='x-large')
fig.colorbar(im_zones, ax=ax5, orientation='vertical', shrink=0.75, pad=0.01, label='Zones', ticks=np.unique(zones))
# Factor of safety
fs_output_file = f"{outputs_dir}/TRfs_min/TRfs_min_{proj_name}_{model}_1.asc"
fs, header = load_ascii_raster(fs_output_file)
im_fs = ax6.matshow(fs, cmap="RdYlGn", alpha=0.7, interpolation="none", vmin=1, vmax=2)
ax6.set_title("FS at $t=0$", fontsize='x-large')
fig.colorbar(im_fs, ax=ax6, orientation='vertical', shrink=0.75, pad=0.01, label='FS')
for ax in [ax0, ax1, ax2, ax3, ax4, ax5, ax6]:
# if ax.get_subplotspec().is_last_col():
if ax == ax6:
ax.xaxis.set_ticks_position('bottom')
ax.yaxis.set_ticks_position('left')
else:
ax.xaxis.set_ticks_position('none')
fig.canvas.header_visible = False
fig.canvas.toolbar_position = 'bottom'
plt.show()
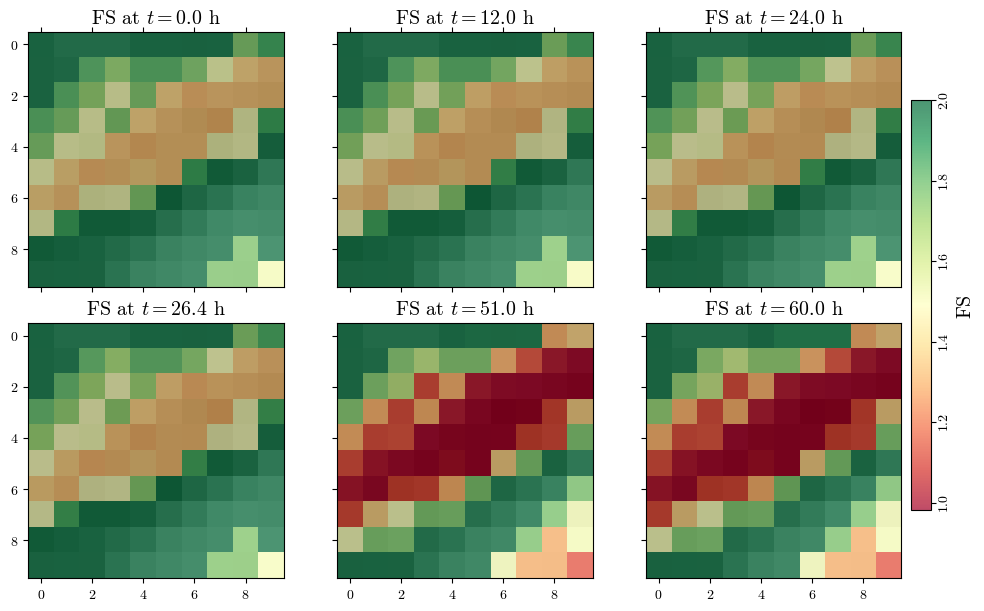
Visualizing spatial outputs of FS#
def read_time(file):
with open(file, 'r') as file:
lines = file.readlines()
time = float(lines[4].split()[-1])
return time
actual_output_times = []
for i in range(1, trg_inputs["n_outputs"]+1):
time = read_time(f'{outputs_dir}/TR_ijz_p_th/TR_ijz_p_th_{proj_name}_{model}_{i}.txt')
actual_output_times.append(time)
actual_output_times, list(output_times)
([0.0, 43200.0, 86400.0, 95040.0, 183600.0, 216000.0],
[0.01, 43200.0, 86400.0, 100800.0, 183600.0, 216000.0])
n_subplots = len(actual_output_times)
nrows=np.ceil(n_subplots/3).astype(int)
fig, axs = plt.subplots(ncols=3, nrows=nrows, figsize=(10, 3*nrows), layout='constrained', sharex=True, sharey=True)
all_fs = []
for i, t in enumerate(actual_output_times):
fs_output_file = f"{outputs_dir}/TRfs_min/TRfs_min_{proj_name}_{model}_{i+1}.asc"
fs, header = load_ascii_raster(fs_output_file)
all_fs.append(fs)
ax = axs[i//3, i%3]
im_hsd = ax.matshow(hsd, cmap="gray", interpolation="none")
im_fs = ax.matshow(fs, cmap='RdYlGn', alpha=0.7)#, vmin=1.0, vmax=2, interpolation="none")
ax.set_title(f"FS at $t={t/3600:.1f}$ h", fontsize='x-large')
# ax.set_xticks([])
# ax.set_yticks([])
min_fs, max_fs = np.nanmin(all_fs), np.nanmax(all_fs) # change vmin and vmax to each subplot
max_fs = min(max_fs, 2)
for ax in axs.flatten():
ax.images[1].set_clim(vmin=min_fs, vmax=max_fs)
ax.xaxis.set_ticks_position('bottom') if ax.get_subplotspec().is_last_row() else None
cbar = fig.colorbar(im_fs, ax=axs, orientation='vertical', shrink=0.75, pad=0.01)
cbar.ax.tick_params(axis="y", labelrotation=90, pad=1.5)
cbar.set_label('FS', rotation=90, size="x-large")
[label.set_va("center") for label in cbar.ax.get_yticklabels()]
fig.canvas.header_visible = False
fig.canvas.toolbar_position = 'bottom'
plt.show()
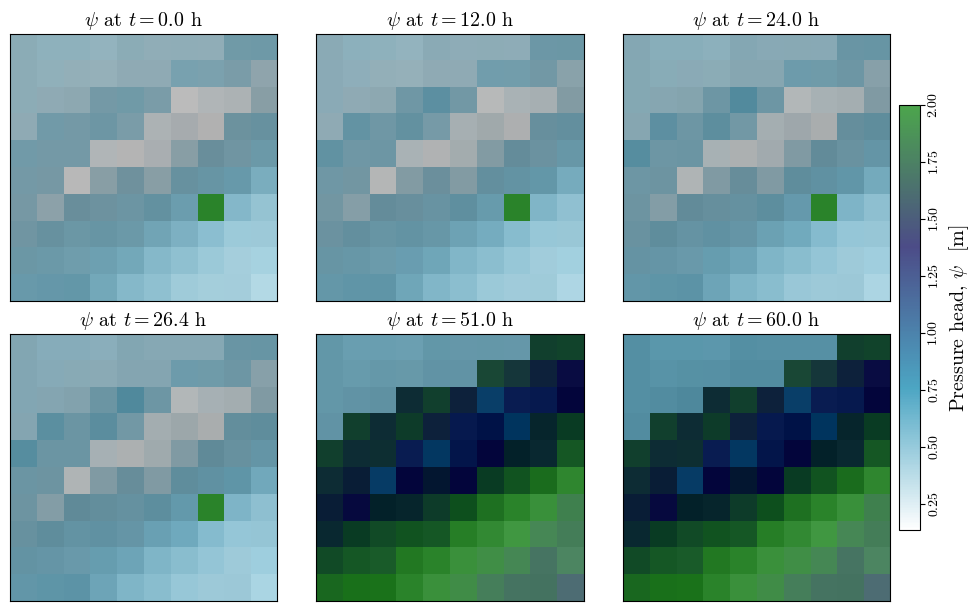
Visualizing spatial outputs of \(\psi\)#
n_subplots = len(actual_output_times)
nrows=np.ceil(n_subplots/3).astype(int)
fig, axs = plt.subplots(ncols=3, nrows=nrows, figsize=(10, 3*nrows), layout='constrained', sharex=True, sharey=True)
all_psi = []
for i, t in enumerate(actual_output_times):
psi_output_file = f"{outputs_dir}/TRp_at_fs_min/TRp_at_fs_min_{proj_name}_{model}_{i+1}.asc"
psi, header = load_ascii_raster(psi_output_file)
all_psi.append(psi)
ax = axs[i//3, i%3]
im_hsd = ax.matshow(hsd, cmap="gray", interpolation="none")
im_psi = ax.matshow(psi, cmap='ocean_r', alpha=0.7, interpolation="none")#, vmin=0, vmax=np.nanmax(depth))
ax.set_title(f"$\psi$ at $t={t/3600:.1f}$ h", fontsize='x-large')
ax.set_xticks([])
ax.set_yticks([])
min_psi, max_psi = np.nanmin(all_psi), np.nanmax(all_psi) # change vmin and vmax to each subplot
for ax in axs.flatten():
ax.images[1].set_clim(vmin=min_psi, vmax=max_psi)
ax.xaxis.set_ticks_position('bottom') if ax.get_subplotspec().is_last_row() else None
cbar = fig.colorbar(im_psi, ax=axs, orientation='vertical', shrink=0.75, pad=0.01)
cbar.ax.tick_params(axis="y", labelrotation=90, pad=1.5)
cbar.set_label('Pressure head, $\psi$ [m]', rotation=90, size="x-large")
[label.set_va("center") for label in cbar.ax.get_yticklabels()]
fig.canvas.header_visible = False
fig.canvas.toolbar_position = 'bottom'
plt.show()
Output to dataframe#
def get_param(zone, param):
return trg_inputs['geoparams'][zone.iloc[0]][param]
def get_df(file):
γ_w = 9.8e3 # [N/m3] unit weight of water
# Reading file
df = pd.read_csv(file, names=['i_tr', 'j_tr', 'elev', 'ψ', 'θ'], skiprows=7, sep='\s+')
# Correcting indices to match numpy notation
df['i'] = df['j_tr'].max() - df['j_tr']
df['j'] = df['i_tr'] - 1
df.drop(columns=['i_tr', 'j_tr'], inplace=True)
# Calculating depths from elevations
elev_max = df.groupby(['i', 'j'])['elev'].transform('max')
df['z'] = elev_max - df['elev'] + 0.001
df = df[['i', 'j', 'elev', 'z', 'ψ', 'θ']]
# Assigning zones and geotechnical parameters
df['zone'] = zones[df['i'], df['j']]
for param_name in trg_inputs['geoparams'][1].keys():
df[param_name] = df.groupby('zone')['zone'].transform(get_param, param=param_name)
# Correting rows with ψ==0 (watertable) due to numerical errors
mask = (df['ψ'] == 0) & (df['z'] > 0.001) & (df['θ'] == df['θ_sat'])
# df.loc[mask, 'z'] = df.loc[mask, 'z'] + 1/df.loc[mask, 'α']
# df.loc[mask, 'elev'] = df.loc[mask, 'elev'] - 1/df.loc[mask, 'α']
# df.sort_values(['i', 'j', 'z'], inplace=True)
df.drop(df[mask].index, inplace=True) # This eliminates the wt rows
# Assigning slope
df['δ'] = slope[df['i'], df['j']]
δrad = np.deg2rad(df['δ'])
# Calculating effective stress parameter, χ
df['χ'] = (df['θ'] - df['θ_res']) / (df['θ_sat'] - df['θ_res'])
# Specific gravity
df['Gs'] = (df['γ_sat']/γ_w - df['θ_sat']) / (1 - df['θ_sat'])
# unit weight at unsat and sat zones
df['γ_nat'] = (df['Gs'] * (1 - df['θ_sat']) + df['θ']) * γ_w
# average unit weight
γ_cumul = df.groupby(['i', 'j'])['γ_nat'].transform('cumsum')
idx = df.groupby(['i', 'j'])['γ_nat'].cumcount() + 1
df['γ_avg'] = γ_cumul / idx
# Factor of safety
fs1 = np.tan(np.deg2rad(df['φ'])) / np.tan(δrad)
fs2_num = df['c'] - df['ψ'] * γ_w * df['χ'] * np.tan(np.deg2rad(df['φ']))
fs2_den = df['γ_avg'] * df['z'] * np.sin(δrad) * np.cos(δrad)
fs = fs1 + fs2_num/fs2_den
fs[fs > 3] = 3
df['fs'] = fs
return df
Verification of FS and head pressure at a point#
cell = (2, 2) # (i, j) indices of the unique cell in the domain (i=y, j=x) in numpy notation
file = f'{outputs_dir}/TR_ijz_p_th/TR_ijz_p_th_{proj_name}_{model}_4.txt'
df = get_df(file)
cell_df = df[(df['i']==cell[0]) & (df['j']==cell[1])]
# cell_df.drop(cell.index[cell['ψ'] == 0], inplace=True)
cell_df.head()
| i | j | elev | z | ψ | θ | zone | c | φ | γ_sat | ... | K_sat | θ_sat | θ_res | α | δ | χ | Gs | γ_nat | γ_avg | fs | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 484 | 2 | 2 | 96.0 | 0.001 | -0.04532 | 0.45 | 1.0 | 3500.0 | 35 | 22000.0 | ... | 1.000000e-07 | 0.45 | 0.05 | -0.5 | 23.0 | 1.0 | 3.263451 | 22000.0 | 22000.0 | 3.0 |
| 485 | 2 | 2 | 95.9 | 0.101 | -0.07715 | 0.45 | 1.0 | 3500.0 | 35 | 22000.0 | ... | 1.000000e-07 | 0.45 | 0.05 | -0.5 | 23.0 | 1.0 | 3.263451 | 22000.0 | 22000.0 | 3.0 |
| 486 | 2 | 2 | 95.8 | 0.201 | -0.10210 | 0.45 | 1.0 | 3500.0 | 35 | 22000.0 | ... | 1.000000e-07 | 0.45 | 0.05 | -0.5 | 23.0 | 1.0 | 3.263451 | 22000.0 | 22000.0 | 3.0 |
| 487 | 2 | 2 | 95.7 | 0.301 | -0.12030 | 0.45 | 1.0 | 3500.0 | 35 | 22000.0 | ... | 1.000000e-07 | 0.45 | 0.05 | -0.5 | 23.0 | 1.0 | 3.263451 | 22000.0 | 22000.0 | 3.0 |
| 488 | 2 | 2 | 95.6 | 0.401 | -0.13190 | 0.45 | 1.0 | 3500.0 | 35 | 22000.0 | ... | 1.000000e-07 | 0.45 | 0.05 | -0.5 | 23.0 | 1.0 | 3.263451 | 22000.0 | 22000.0 | 3.0 |
5 rows × 21 columns
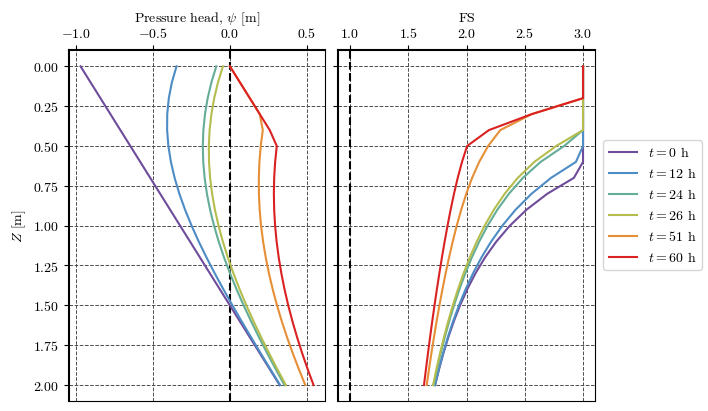
Plotting results at a point#
clrs = ['#6F4C9B', '#6059A9', '#5568B8', '#4E79C5', '#4D8AC6', '#4E96BC',
'#549EB3', '#59A5A9', '#60AB9E', '#69B190', '#77B77D', '#8CBC68',
'#A6BE54', '#BEBC48', '#D1B541', '#DDAA3C', '#E49C39', '#E78C35',
'#E67932', '#E4632D', '#DF4828', '#DA2222']
cmap = mpl.colors.LinearSegmentedColormap.from_list('cmap_name', clrs)
# cmap
fig, axs = plt.subplots(nrows=1, ncols=2, figsize=(7, 4), sharey=True, layout='constrained')
palette = mpl.colormaps['gnuplot_r']
palette = cmap
for ax in axs:
ax.grid(True, color='0.3', ls='--', lw=0.7)
ax.spines["top"].set_linewidth(1.5)
ax.spines["left"].set_linewidth(1.5)
ax.xaxis.set_ticks_position('top')
ax.xaxis.set_tick_params(top=True, bottom=False)
ax.xaxis.set_label_position('top')
ax.set_prop_cycle('color', palette(np.linspace(0, 1, len(output_times))))
axs[0].axvline(0, color='k', lw=1.5, ls='--')
axs[1].axvline(1, color='k', lw=1.5, ls='--')
for n in np.arange(len(output_times)) + 1:
file = f'TRIGRS_2/outputs/TR_ijz_p_th/TR_ijz_p_th_{proj_name}_{model}_{n}.txt'
t = read_time(file)
df = get_df(file)
df_cell = df[(df['i']==cell[0]) & (df['j']==cell[1])]
axs[0].plot(df_cell['ψ'], df_cell['z'], label=f'$t={t/3600:.0f}$ h')
axs[1].plot(df_cell['fs'], df_cell['z'], label=f'$t={t/3600:.0f}$ h')
axs[0].set(xlabel='Pressure head, $\\psi$ [m]', ylabel='$Z$ [m]')
axs[1].set(xlabel='FS')
axs[1].invert_yaxis()
handles, labels = axs[0].get_legend_handles_labels()
fig.legend(handles, labels, loc='outside right center')
fig.canvas.header_visible = False
fig.canvas.toolbar_position = 'bottom'
plt.show()